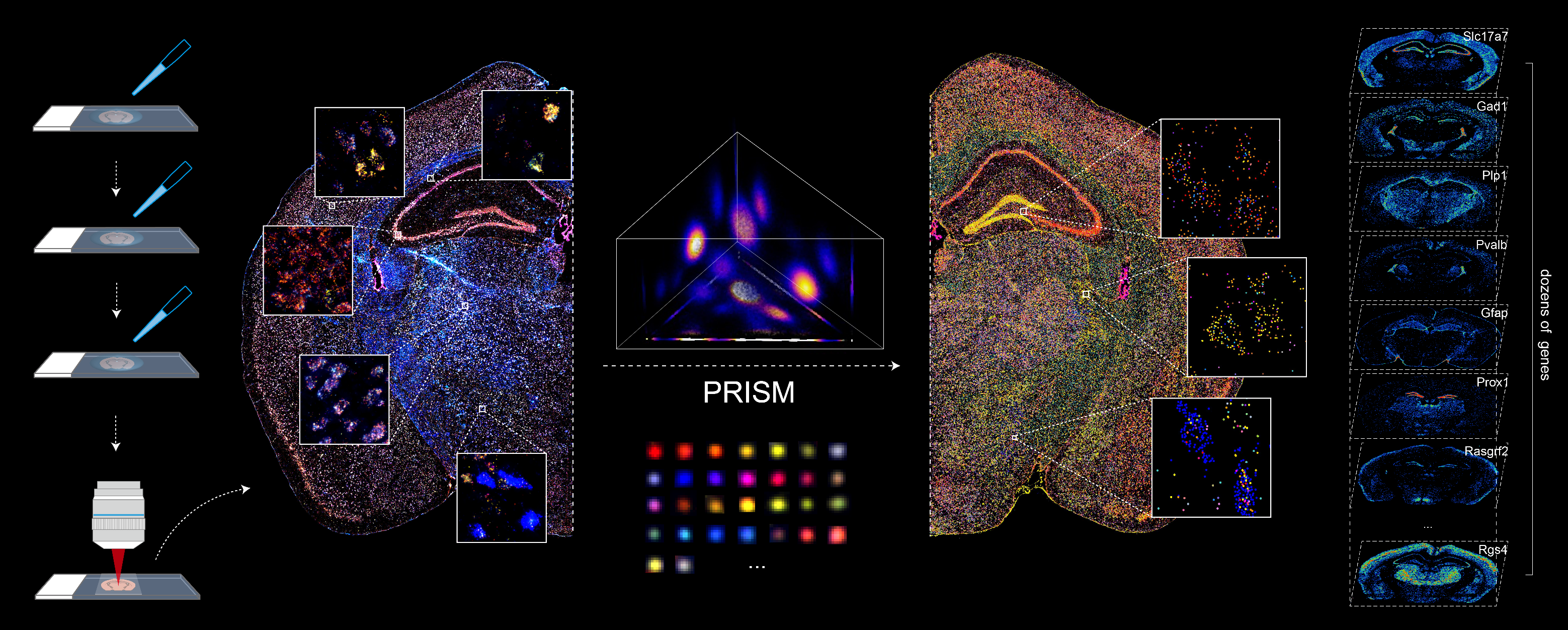
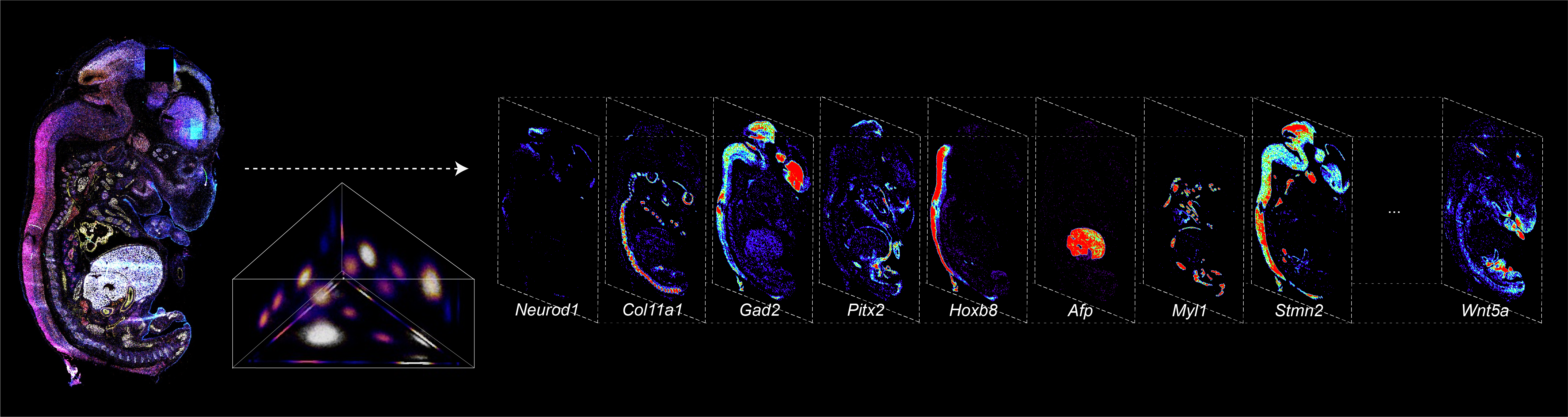
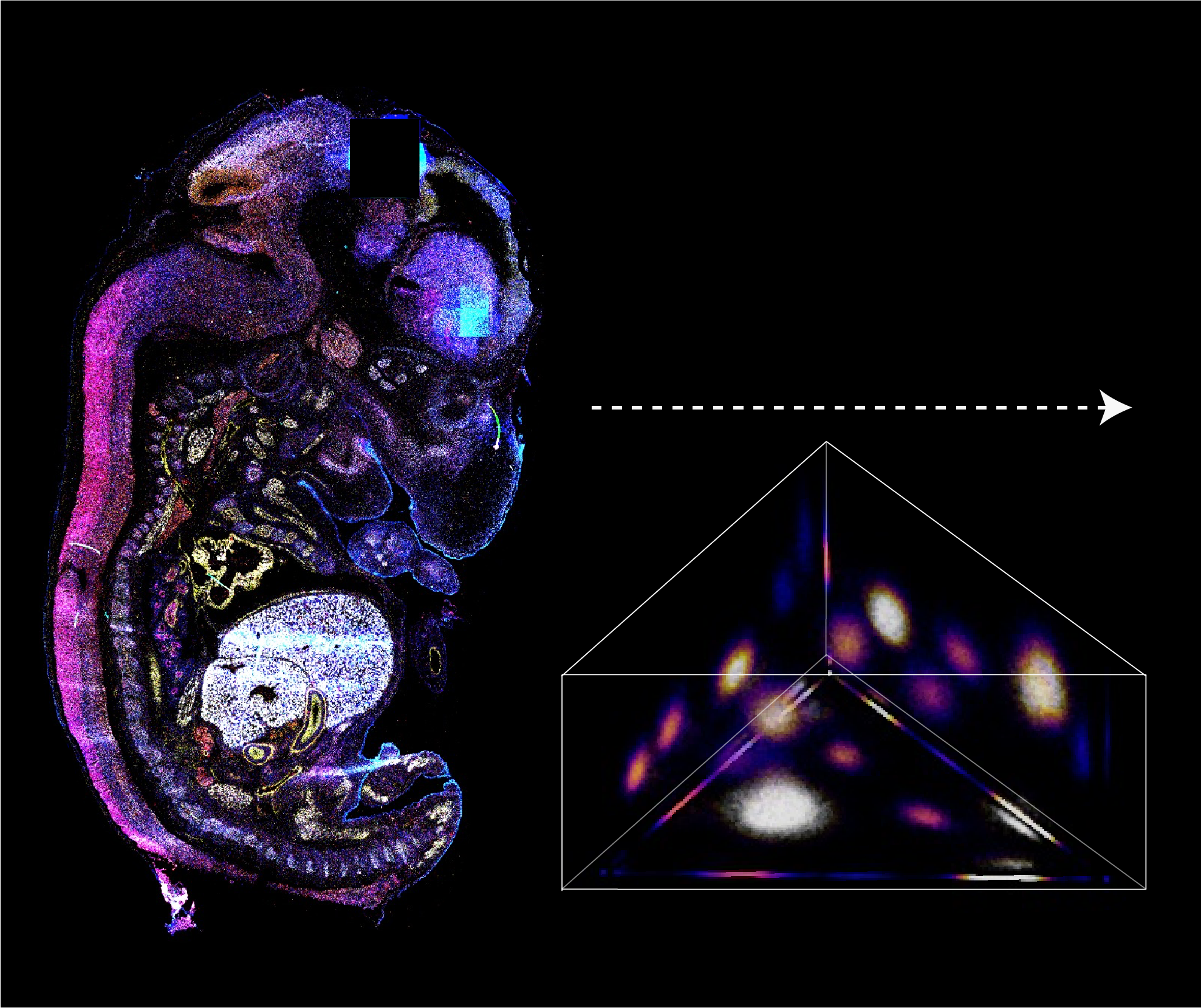
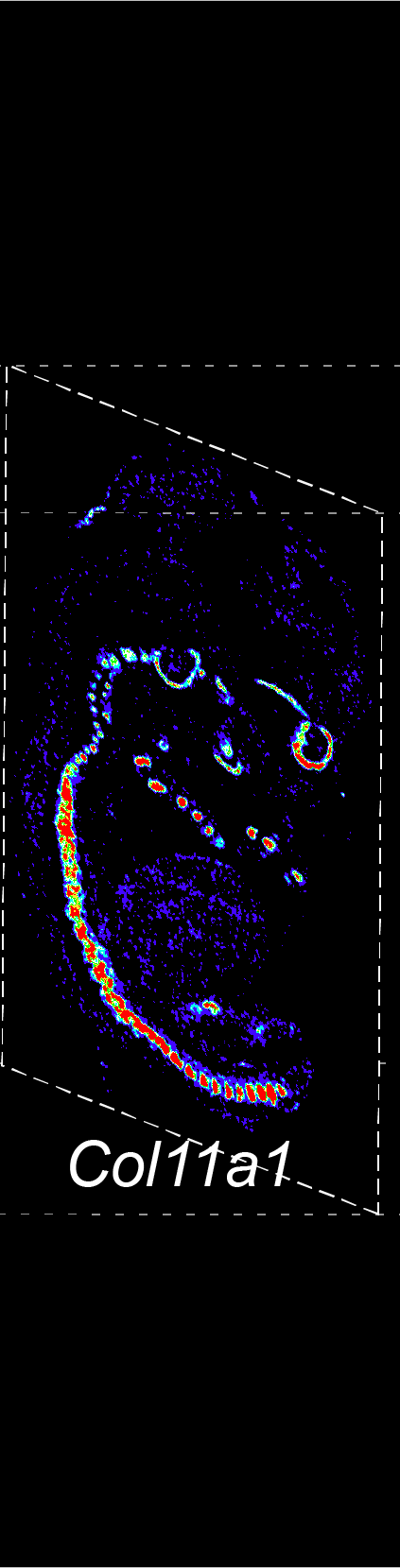
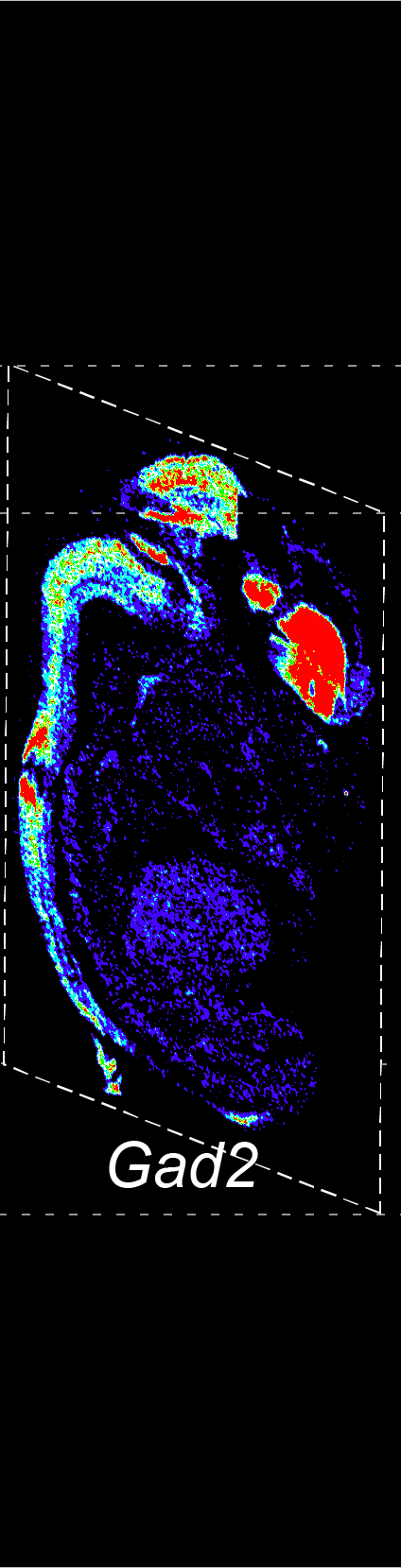
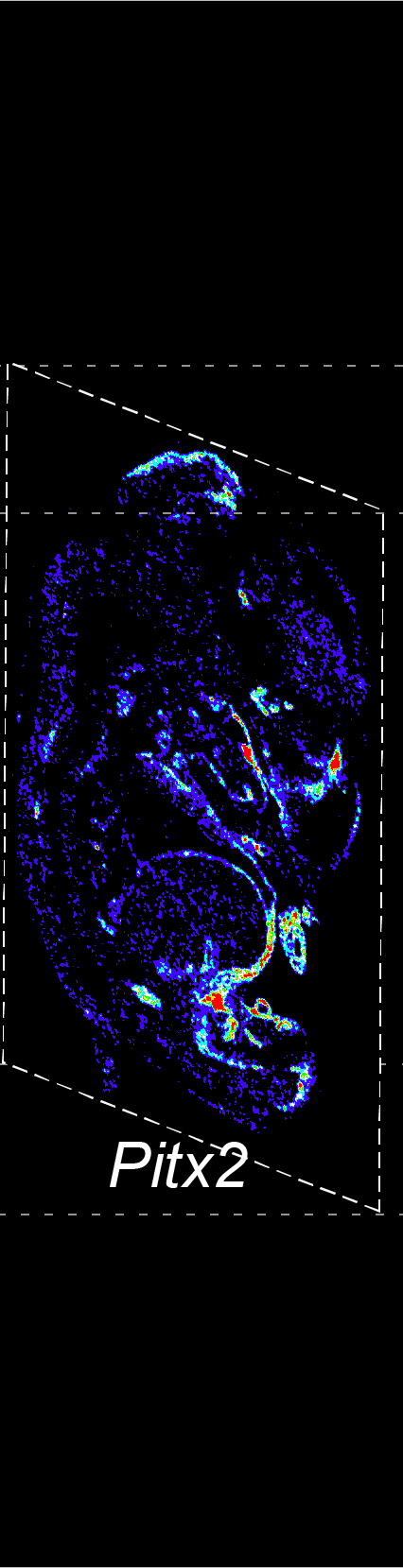
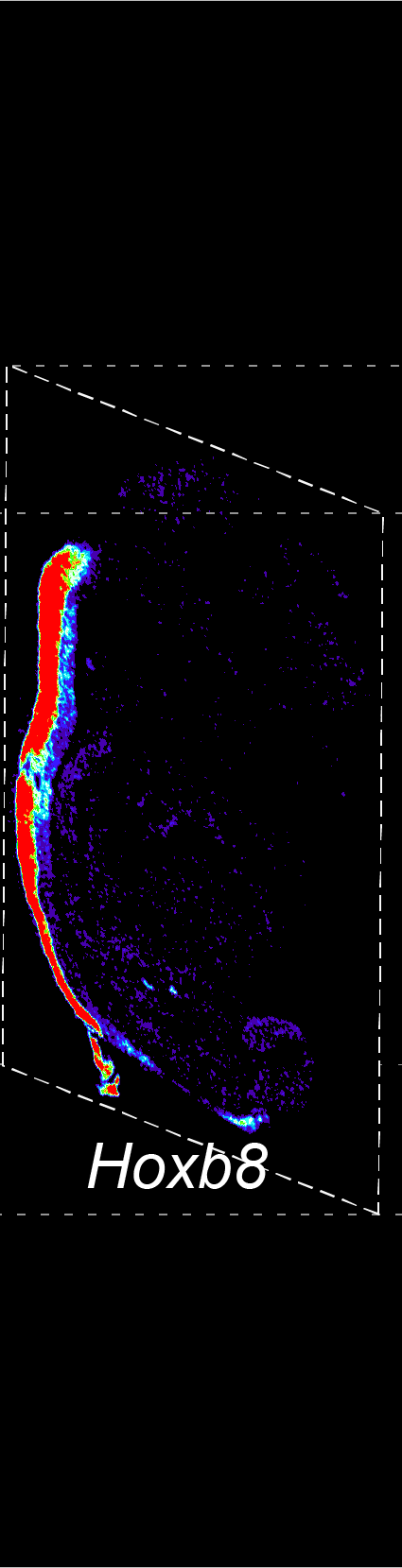
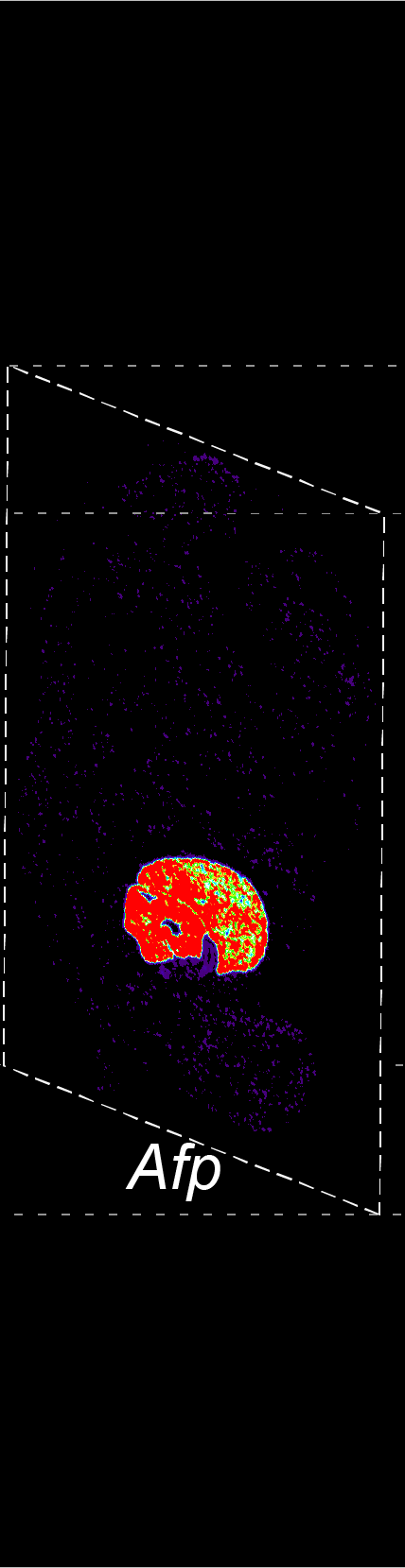
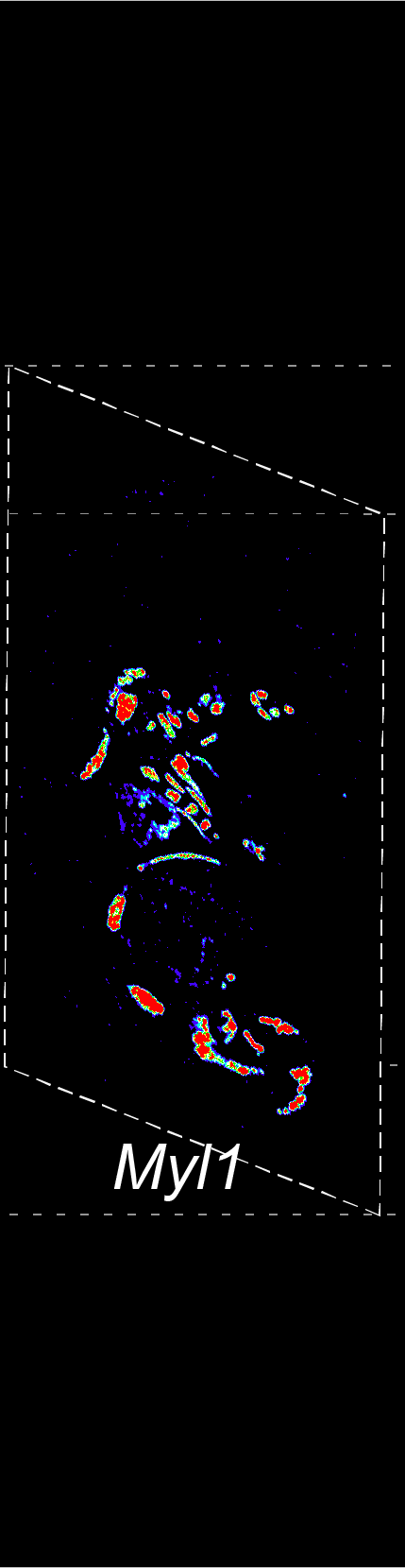
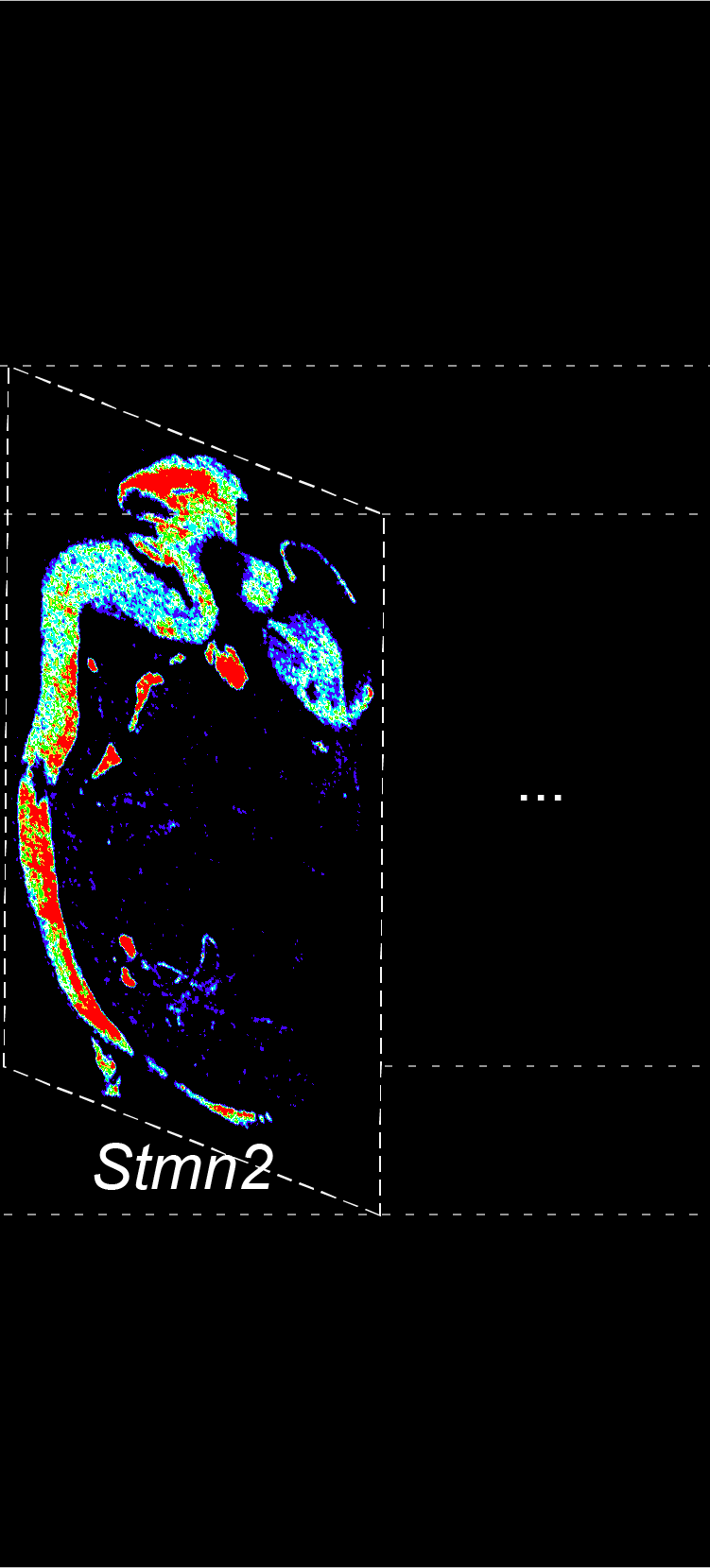
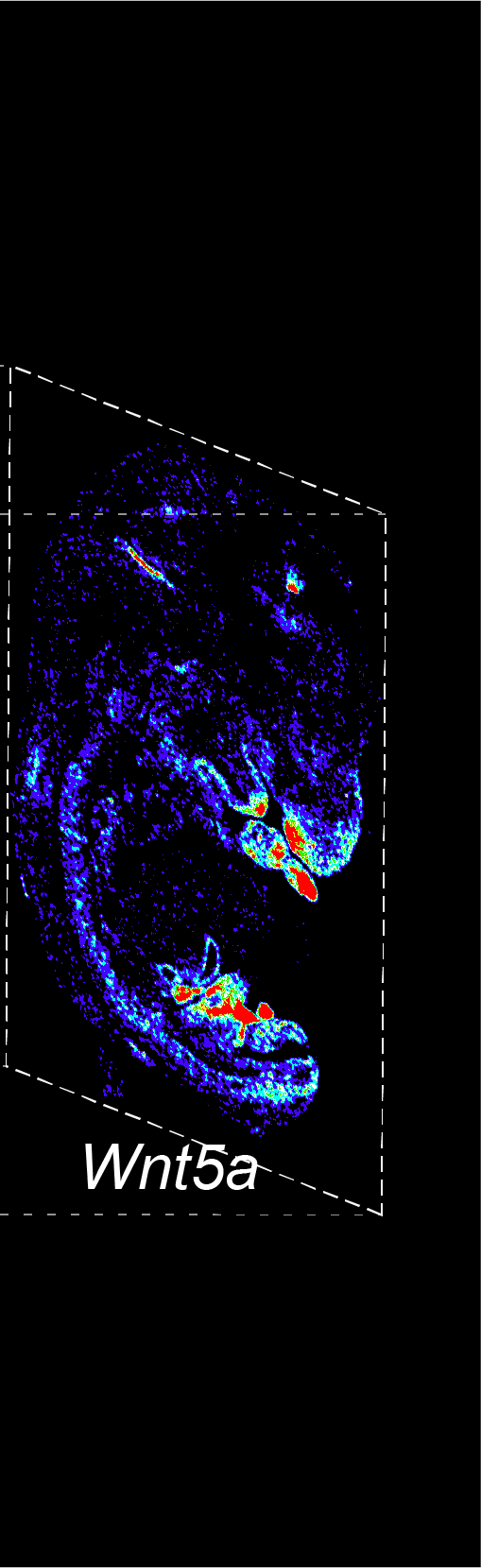
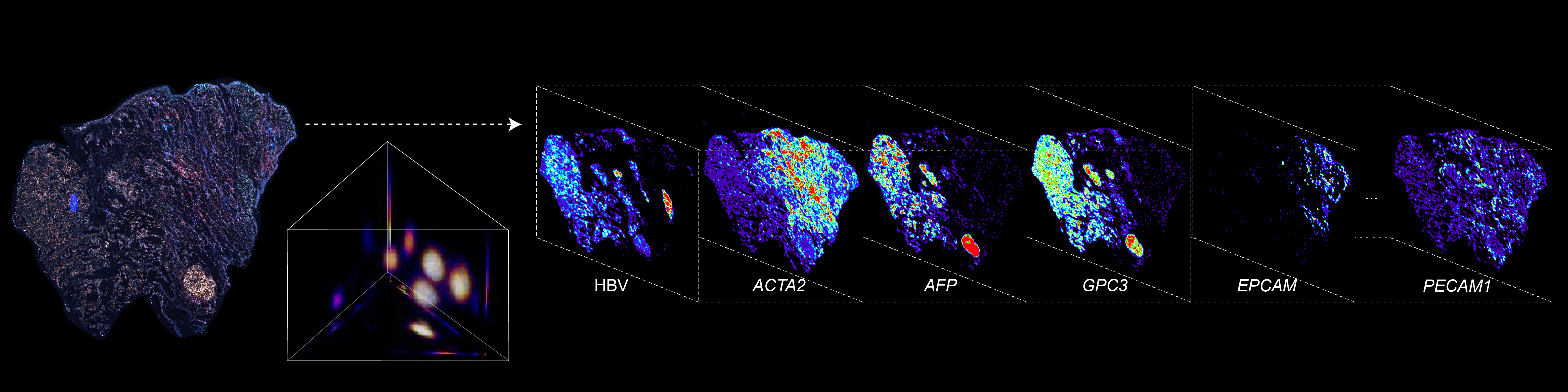
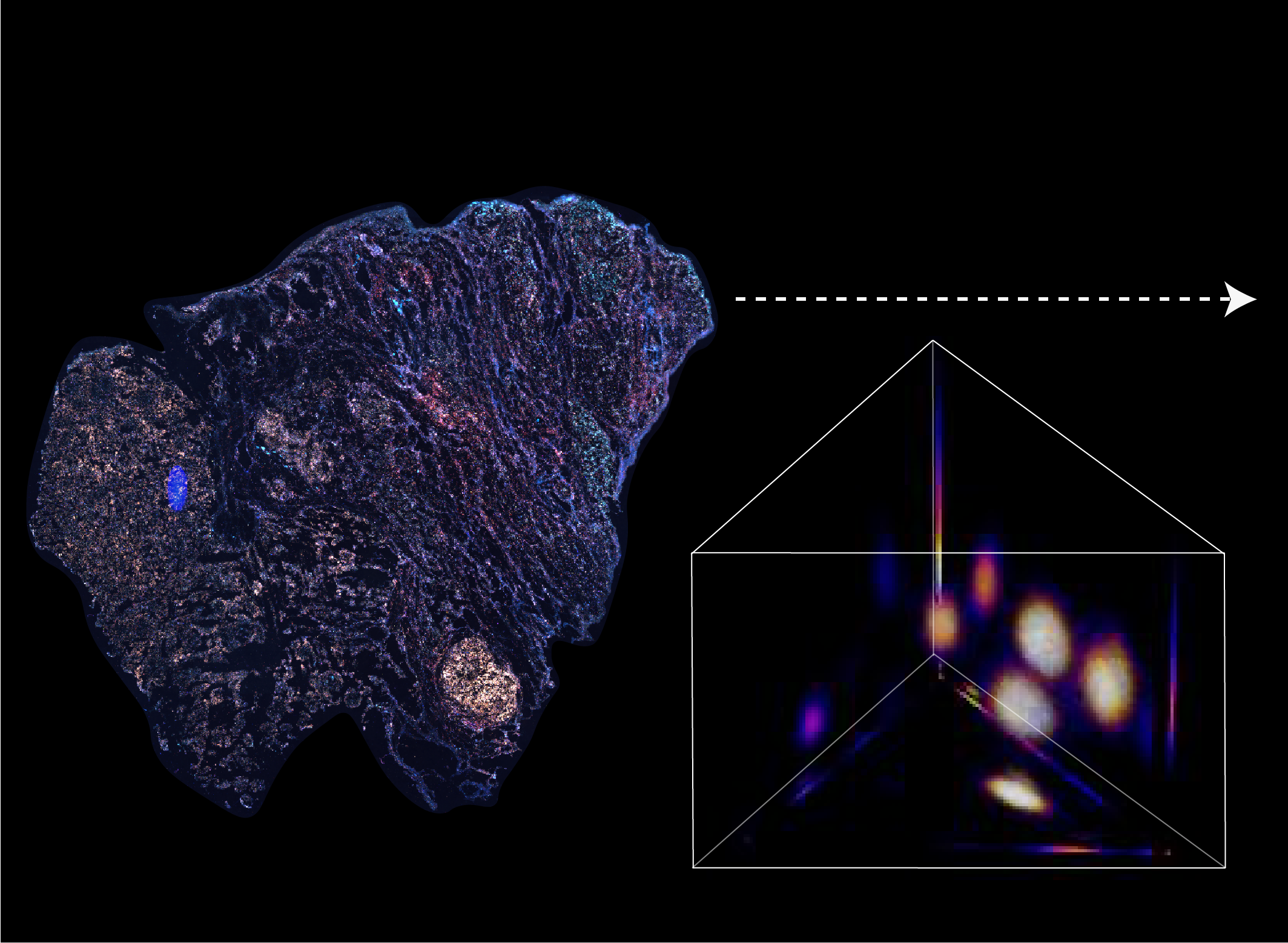
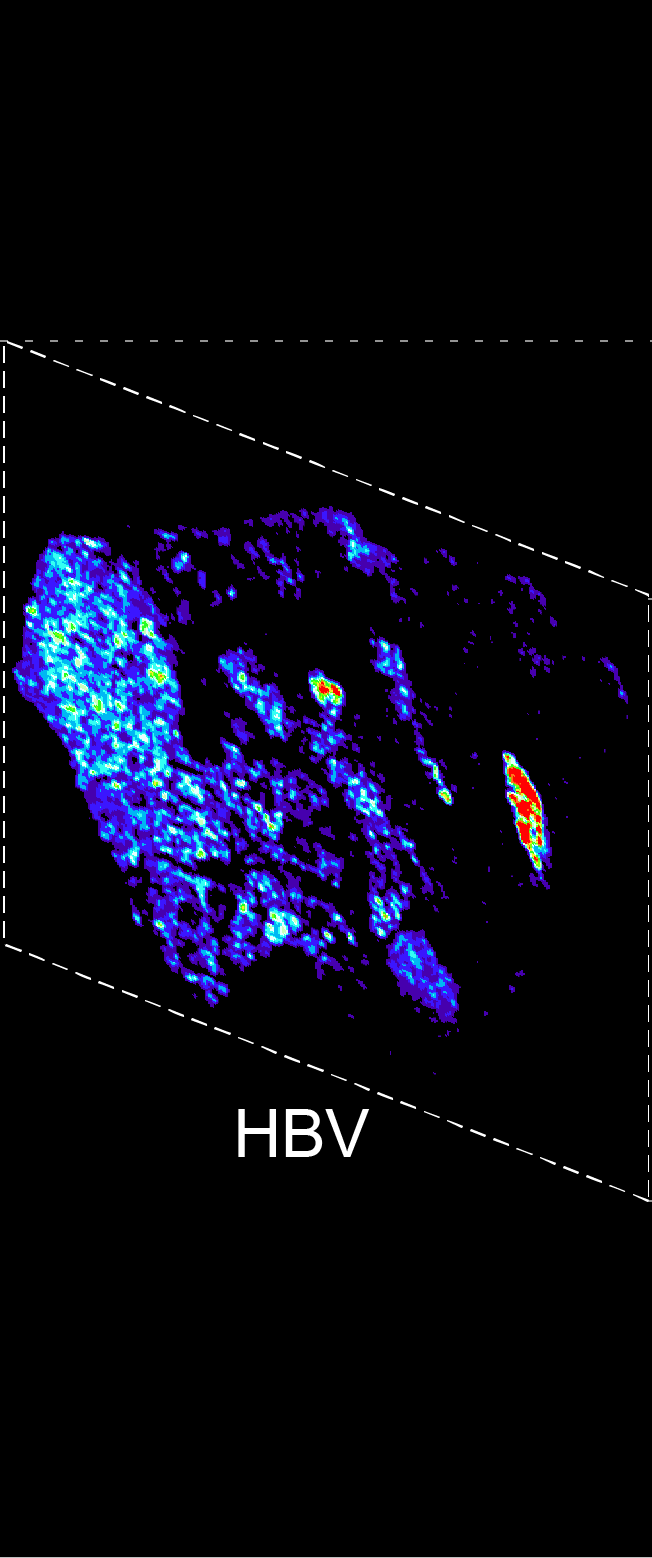
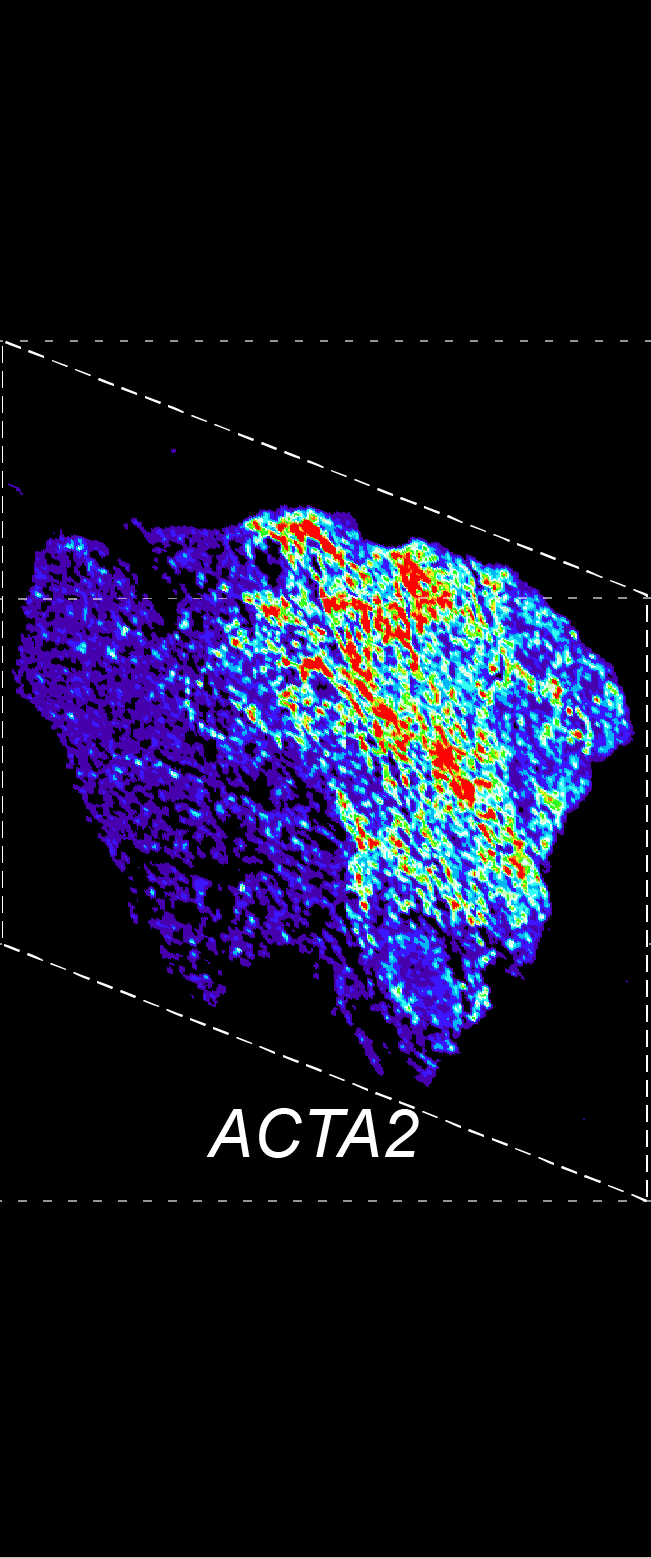
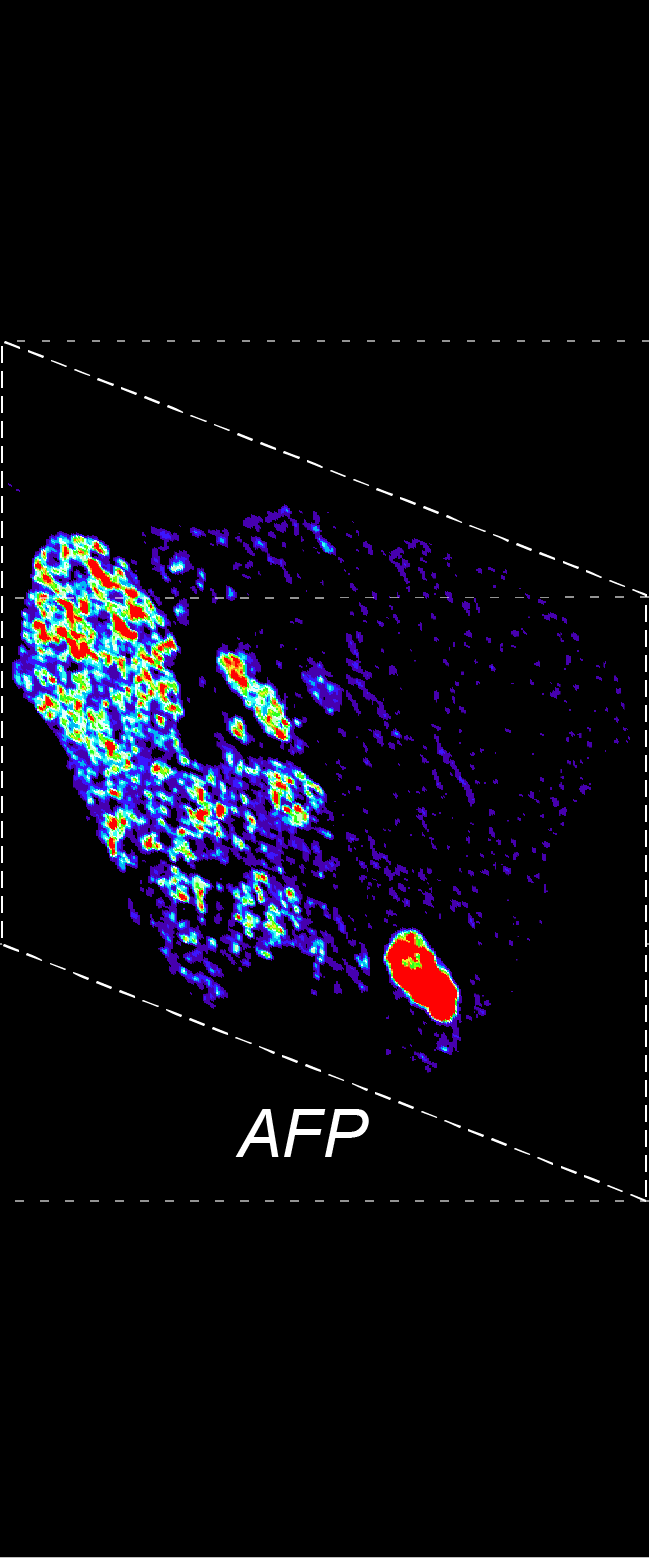
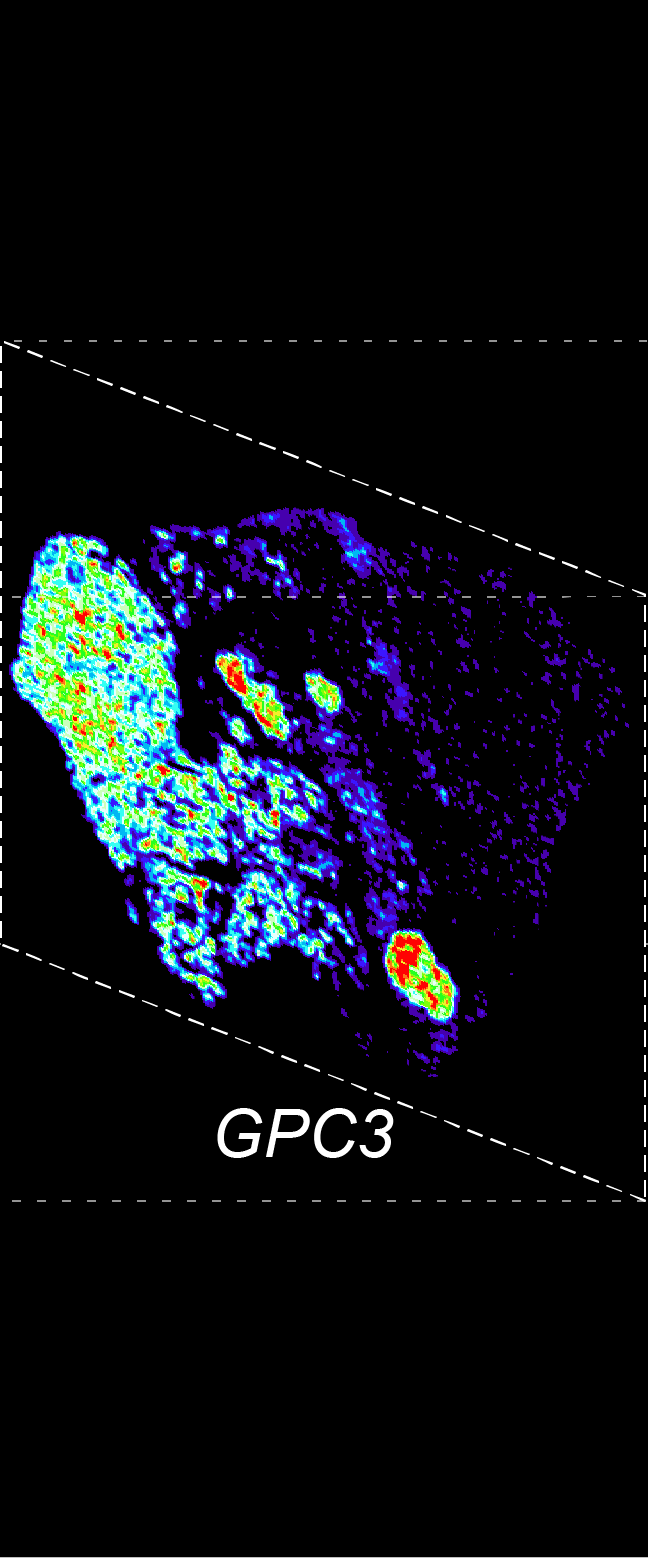
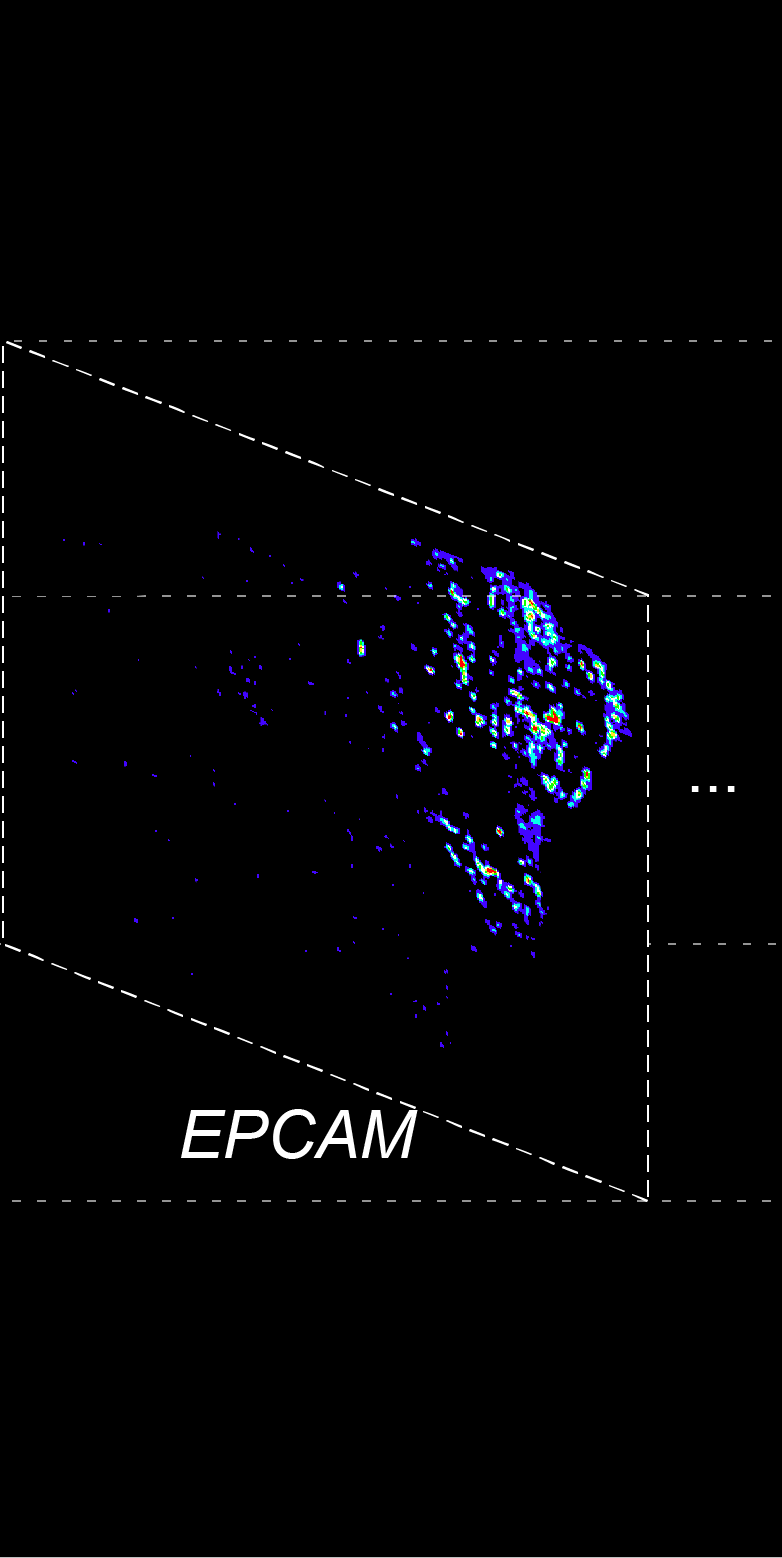
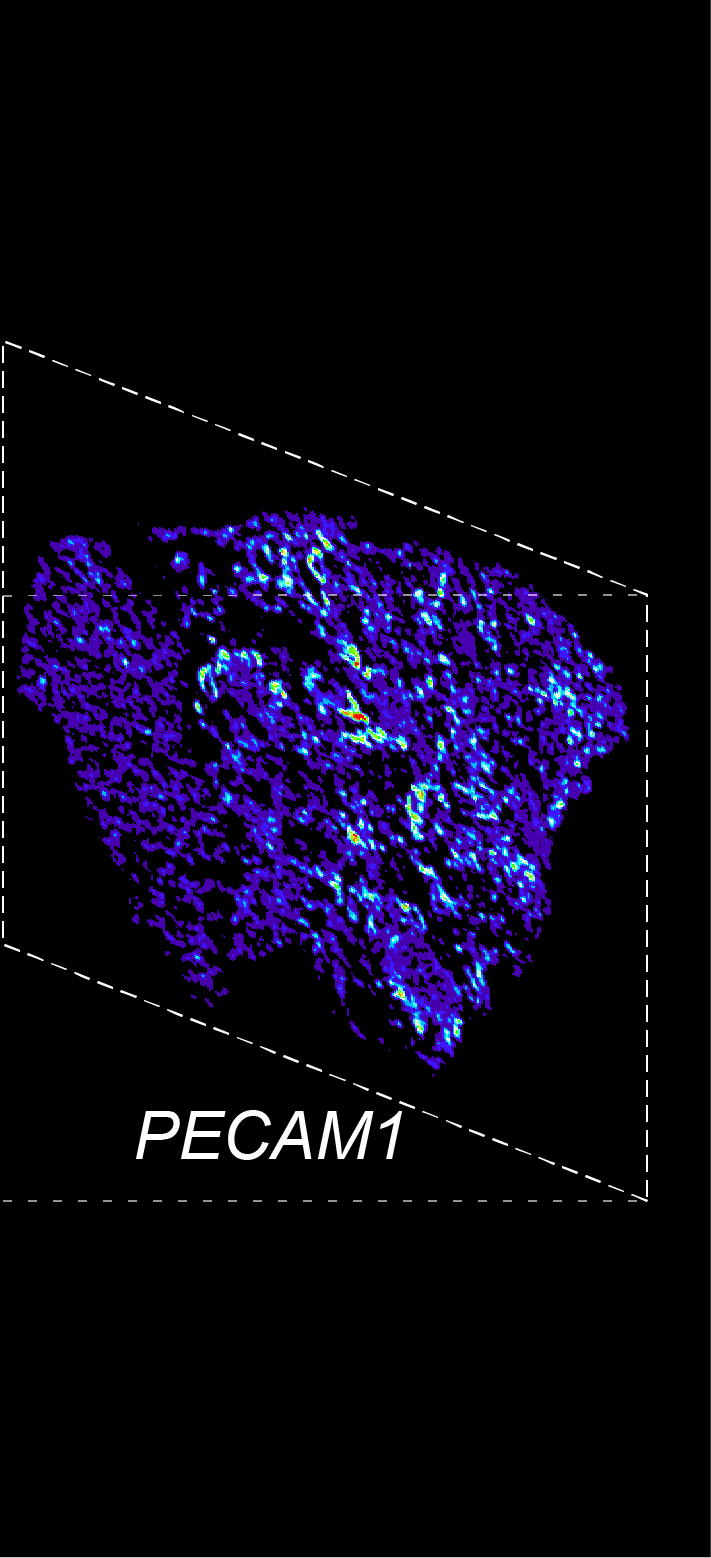
BARCODE EXPANSION
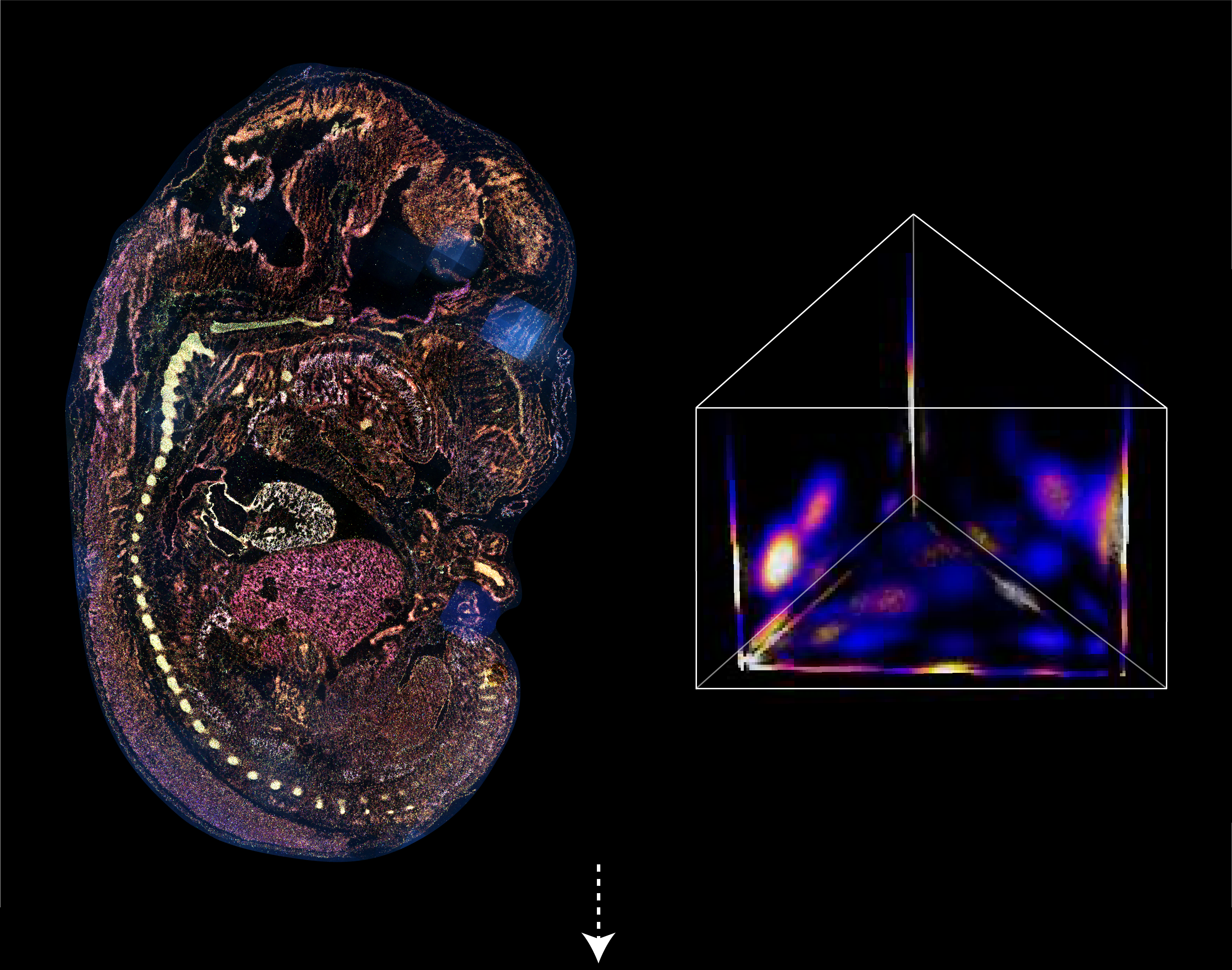
Barcoded padlock probes are designed to hybridize targeted gene transcripts and are amplified locally into clones via rolling circle amplification. Each barcode contains segments corresponding to a spectral channel, taking the spectral combination (both wavelength and intensity) for coding.
The intensity information was encoded in the ratio between fluorescently labeled and unlabeled secondary probes. Using four channels, we have experimentally verified the barcode multiplexity of up to 64 genes in a single imaging round.
CODING PRINCIPLE
Variations in the sizes of amplified nanoballs could make specific codes indistinguishable in spectral color space. To better understand the discriminability of barcodes, we introduced the concept of ‘radius vector’ in color space, and only angularly distinct barcode vectors were considered legitimate.
To ensure ample dispersion of barcode vectors, we chose the plane (Ch1+Ch2+Ch3 = 4) in the coding space as the constraint, reaching an optimal differentiability of intersected barcodes by calculating the angles between any two barcode vectors and eliminating the crosstalk between barcodes.